ARIA API Documentation
This document intends on providing all of the information required to establish a connection with the Instruct proposals submission system and import data into a local system. Code examples are provided where possible, and it is strongly advised that you follow through the demo using the provided credentials. The demo provides an example of just one potential workflow of the API, however your own implementation does not have to follow the same workflow.
OAuth Documentation
OAuth endpoints are in place to establish per-user access tokens. To get a client_id and a client_secret contact Instruct to discuss your requirements. Each client_id has a limited scope of access based on your requirement and must not be shared between different software implementations. All OAuth responses are received in JSON format, however the ARIA output endpoints can be handled with multiple output formats.
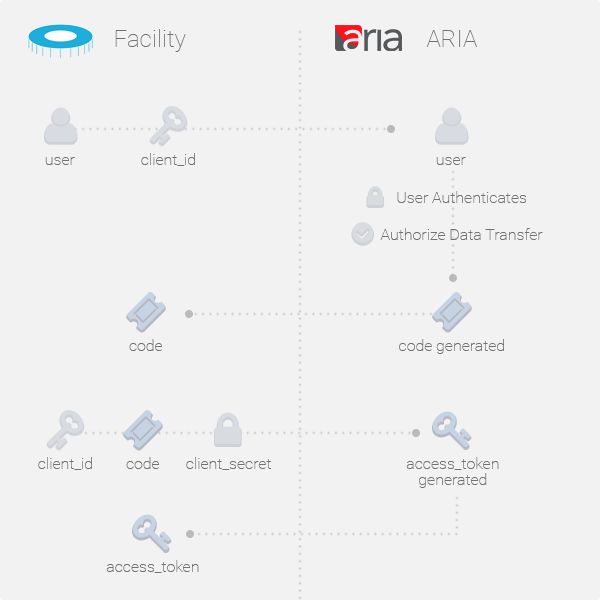
Before continuing with OAuth information
Make sure you have gone through the demo before continuing with this section of the API documentation.
OAuth Credentials
You will need to have client ID and secret provided by Instruct. These cannot be generated by developers and must be provided by Instruct. Contact the Instruct development team to get your keys. Testing OAuth credentials are provided at the end of this API section, which have access only to the test proposal submitted by the test user (who's credentials are also available at the end of this document).
| OAuth Service | Endpoint URI | Description |
|---|---|---|
| authorize | https://www.structuralbiology/ws/oauth/authorize | To generate a user authorization token. |
| token | https://www.structuralbiology/ws/oauth/token | To generate an access token or refresh an existing token. |
| response | https://www.structuralbiology/ws/oauth/response | Data output endpoint accessible with an access token. |
/authorize
Initiation of the authorization can be done with either a form or URL pointed towards the authorization endpoint URI with the required HTTP variables described below.
| Variable | Values | Required | Description |
|---|---|---|---|
| client_id | your client_id string | required | To generate a user authorization token. |
| state | a unique per-session string | required | The state is used to prevent malicious use of . |
| response_type | code | required | Define the name of the variable returned to you with the authorization code. |
| force_login | false | optional | You can optionally force the user to re-authenticate when they have an active single sign-on session. |
| redirect_url | url | optional | This is where the authorization request will be redirected. If omitted the user will be redirected to the configured end-point for your client_id. |
| scope | proposallist%20proposal | optional | A list of services you want to authorize with the user. If omitted this will default to all endpoints associated with your client_id. |
<a href="https://www.structuralbiology.eu/ws/oauth/authorize?client_id=4Fgp5SLGRqxaVL5Z534Gou7bjoTjhbyFc3qaoZg5k865GXvEoTG8je4PtMVYKo6m&state=changeme&response_type=code">Import Proposal from ARIA</a>
/token
To receive an access token you will need to request from the OAuth token service. This must be constructed as an HTTPS POST request from your server side and not from the client side as the request includes your client_secret token.
| Variable | Values | Required | Description |
|---|---|---|---|
| client_id | your client_id string | required | Your client_id for your service. |
| client_secret | your client_secret string | required | Your client_secret for your service. |
| grant_type | authorization_code refresh_token |
required | Define what request type is being initiated. The majority of requests will be refresh_tokens. |
| code | The authorization code recieved from authorization service. | dependent | This is required for authorization_code grant_type. |
| refresh_token | Client refresh token. | dependent | This is required for refresh_token grant_type. |
// The following example is getting an access token from a successful authorization code
// The following example is getting an access token from a successful authorization code
if(isset($_GET['code']) && isset($_GET['state'])){
$code = $_GET['code'];
$state = $_GET['state'];
// quickly sanitize input
if(!preg_match('/[A-Za-z0-9]+/', $code))
exit('Authentication code invalid');
if(!preg_match('/[A-Za-z0-9]+/', $state))
exit('State is invalid');
// verify the state before continuing on
if($state != $_SESSION['OAstate'])
exit('State is invalid');
// manually build OAuth packet - this can be replaced for an off-shelf solution
$OApacket = array(
'grant_type' => 'authorization_code',
'code' => urlencode($code)
);
// start curl connection
$ch = curl_init();
curl_setopt( $ch, CURLOPT_URL, $OAservice.'token' );
curl_setopt( $ch, CURLOPT_FOLLOWLOCATION, true );
curl_setopt( $ch, CURLOPT_RETURNTRANSFER, true );
curl_setopt( $ch, CURLOPT_AUTOREFERER, true );
curl_setopt( $ch, CURLOPT_SSL_VERIFYPEER, false );
curl_setopt( $ch, CURLOPT_MAXREDIRS, 10 );
// pass clientID and clientSecret
curl_setopt( $ch, CURLOPT_USERPWD, $client_id .':'. $client_secret );
// add OAuth packet and define HTTP POST
curl_setopt( $ch, CURLOPT_POST, true );
curl_setopt( $ch, CURLOPT_POSTFIELDS, http_build_query($OApacket) );
// run curl and capture output
if(!$content = curl_exec( $ch ))
exit('Could not fetch access token');
curl_close ( $ch );
// convert JSON into object
if(!$authorization = json_decode($content))
exit('JSON parsing error');
if($authorization->access_token){
// we will store access tokens as cookies for this demo, but you will want to store them somewhere more persistent (database)
setcookie('access_token', $authorization->access_token, time()+$authorization->expires_in);
// this refresh token actually has an expiry of 28 days, however for the demo it's useful to expire within a single day
setcookie('refresh_token', $authorization->refresh_token, time()+60*60*24*1);
$_COOKIE['access_token'] = $authorization->access_token;
}
}
/proposallist
| Variable | Values | Required | Description |
|---|---|---|---|
| access_token | access token | required | The user's access token string. |
| aria_reponse_format | json xml |
required | What format you require the returned data in. |
$.ajax({
url: 'https://www.structuralbiology.eu/ws/oauth/proposallist',
dataType: 'json',
method: 'POST',
data: {
access_token: access_token,
aria_response_format: 'json'
},
success: function(data){
if(!data.error){
// do something with the proposal list data
} else
alert(data.error+'\n'+data.error_description);
}
});
/proposal
| Variable | Values | Required | Description |
|---|---|---|---|
| access_token | access token | required | The user's access token string. |
| aria_reponse_format | json xml |
required | What format you require the returned data in. |
| aria_pid | integer | required | Integer ID of the proposal you are accessing. |
$.ajax({
url: 'https://www.structuralbiology.eu/ws/oauth/proposal',
dataType: 'json',
method: 'POST',
data: {
access_token: access_token,
aria_response_format: 'json',
aria_pid: $(this).data('pid')
},
success: function(data){
if(!data.error){
// do something with the proposal data
} else
alert(data.error+'\n'+data.error_description);
}
});
Demo connection details
The following client connection details have access to the test data that is used in the API demo. No additional data can be accessed using these tokens and you will need to have your own tokens for your own data.
| ID | Key |
|---|---|
| client_id | 4Fgp5SLGRqxaVL5Z534Gou7bjoTjhbyFc3qaoZg5k865GXvEoTG8je4PtMVYKo6m |
| client_secret | oLAQPIiKYqLPSoQhmCcPnEPf5GQqJIbtTbfafKk83rKk62OHZmXIRhjon14sA7EP |
API Keys are being maintained as a legacy connection format. Should the demand for them return they will be documented here.
Proposal List
| Output | Format | Description |
|---|---|---|
| pid | integer | A static identifier for the proposal. You should store this against any resulting proposal created in your local system. |
| title | text | |
| statusID | integer | A full matrix of each status ID is available in the FAQ section. |
| status | text | A textual version of the proposal status that can be used to display to the user. |
| submitted | unix timestamp | |
| display | text | This contains a pre-formatted display of the proposal. It is recommended this is used when displaying the proposal to the user for consistency and usability. |
{
"proposals": [
{
"pid": "1582",
"title": "API Test with Crystallisation and MX beamtime",
"statusID": "5",
"status": "Approved",
"submitted": "1454583055",
"display": "PID: 1582 - API Test with Crystallisation and MX beamtime"
}
]
}
Proposal
| Output | Format | Description |
|---|---|---|
| pid | integer | A static identifier for the proposal. You should store this against any resulting proposal created in your local system. |
| title | text | |
| statusID | integer | A full matrix of each status ID is available in the FAQ section. |
| status | text | A textual version of the proposal status that can be used to display to the user. |
| submitted | unix timestamp | |
| display | text | This contains a pre-formatted display of the proposal. It is recommended this is used when displaying the proposal to the user for consistency and usability. |
{
"proposal": {
"pid": "1582",
"title": "API Test with Crystallisation and MX beamtime",
"statusID": "5",
"status": "Approved",
"submitted": "1454583055",
"owner": {
"firstName": "ARIA",
"lastName": "Test",
"email": "aria@structuralbiology.eu",
"country": "gb",
"organisation": "Instruct",
"sso": [{
"idp": "https:\/\/www.structuralbiology.eu\/idp\/shibboleth",
"id": "aria.test@structuralbiology.eu"
}
]
},
"pi": {
"firstName": "ARIA",
"lastName": "Test",
"email": "aria@structuralbiology.eu",
"country": "gb",
"organisation": "Instruct",
"sso": [{
"idp": "https:\/\/www.structuralbiology.eu\/idp\/shibboleth",
"id": "aria.test@structuralbiology.eu"
}
]
},
"team": [{
"firstName": "Callum",
"lastName": "Smith",
"email": "callum@strubi.ox.ac.uk",
"country": "gb",
"organisation": "University of Oxford",
"sso": [{
"idp": "https:\/\/www.structuralbiology.eu\/idp\/shibboleth",
"id": "callum@structuralbiology.eu"
}, {
"idp": "https:\/\/umbrellaid.org\/idp\/shibboleth",
"id": "https:\/\/umbrellaid.org\/idp\/shibboleth!https:\/\/www.structuralbiology.eu\/shibboleth!0f9fd086-94ad-4d59-bb66-05f15d95691f"
}, {
"idp": "https:\/\/umbrellaid.org\/idp\/shibboleth",
"id": "0f9fd086-94ad-4d59-bb66-05f15d95691f"
}, {
"idp": "https:\/\/registry.shibboleth.ox.ac.uk\/idp",
"id": "whgu0537@ox.ac.uk"
}
]
}
],
"fields": {
"11": {
"data": "This proposal is available as a test for the demo of the OAuth automated proposal sharing method. The rest of the content will be filler text.\r\n\r\nLorem ipsum dolor sit amet, consectetur adipiscing elit. Suspendisse blandit justo justo, vitae iaculis est molestie eget. Sed pellentesque hendrerit leo a mollis. In eget libero purus. Cras rutrum faucibus eleifend. Proin suscipit condimentum lacus quis faucibus. Pellentesque vel velit eu orci condimentum facilisis ut ac felis. Mauris ut purus vitae sem convallis dapibus eget ut leo. Nunc urna diam, elementum sed molestie id, semper non purus.\r\n\r\nSed sed ex erat. Aliquam scelerisque luctus nunc, sit amet tempor elit convallis ut. Pellentesque ipsum ligula, hendrerit a felis quis, condimentum condimentum lacus. Morbi tincidunt ullamcorper elit, eget iaculis est semper quis. Proin sit amet odio at mi consequat feugiat non id ipsum. Proin faucibus dignissim ligula, non semper orci aliquam non. Quisque bibendum vitae erat ac maximus. Morbi et velit convallis arcu gravida vehicula nec ac lacus. Aliquam sed lacus id enim porttitor iaculis. Phasellus rhoncus consequat accumsan.\r\n\r\nClass aptent taciti sociosqu ad litora torquent per conubia nostra, per inceptos himenaeos. Donec consequat diam nec est faucibus posuere. Praesent eget sodales nisl, nec interdum dui. Integer vel fermentum velit. Donec et orci diam. Donec vel enim sed odio facilisis vehicula eget id lacus. Maecenas rhoncus sagittis arcu eget mollis. Vivamus gravida nulla in ultrices pretium. Maecenas rutrum eget metus eget auctor. Donec sodales vitae sapien nec maximus. Integer lobortis id metus at laoreet. Maecenas pretium vulputate ligula, ac accumsan sapien pharetra at. Ut convallis egestas bibendum.\r\n\r\nNam ullamcorper lacus massa, placerat luctus lorem aliquet ut. Praesent ac felis risus. Vivamus id euismod nisl, et tempor leo. In ac lacus maximus enim euismod ullamcorper. Cras efficitur neque feugiat, ornare mauris non, iaculis tortor. Mauris est leo, lacinia sit amet ultrices quis, faucibus nec mauris. Aliquam ultrices tincidunt odio, sit amet pellentesque ex posuere in.\r\n\r\nDuis a porta lacus. Integer semper turpis justo, laoreet consectetur nunc tempus non. Suspendisse eget neque leo. Vestibulum iaculis enim turpis, vitae sagittis purus tempus quis. Nulla ut euismod diam. Nunc nec ornare nibh. In et lacus imperdiet nibh pellentesque dictum nec id orci. Quisque bibendum mi vel lacus laoreet faucibus.",
"title": "Scientific background, significance and objectives",
"type": "2"
},
"12": {
"data": "The research programme will consist of:\r\n\r\nMorbi dapibus nec turpis at gravida. Sed pulvinar sollicitudin velit, ac scelerisque nisl laoreet ut. Aenean malesuada, sapien nec finibus dignissim, sem justo ultrices enim, ut rhoncus enim magna vel sapien. Quisque dignissim massa nibh, et mollis sem cursus et. Nam efficitur dictum odio. Vivamus viverra nisi eget nunc scelerisque laoreet. Vestibulum ante ipsum primis in faucibus orci luctus et ultrices posuere cubilia Curae; Duis justo sem, interdum nec egestas vehicula, gravida eu nibh. Duis placerat eros nec dolor elementum feugiat. Pellentesque varius orci id tortor lobortis, vel feugiat justo ornare. Vestibulum eget imperdiet diam. Nulla facilisi. Curabitur quis ipsum vel ante varius vestibulum. Fusce ornare, augue quis pulvinar posuere, nisi libero efficitur nisl, vel congue ipsum nisi non ante.\r\n\r\nFusce consequat maximus orci, ut tempus libero ultrices vitae. Ut turpis augue, varius et efficitur non, ultrices non mi. Maecenas nec urna sed mi auctor malesuada. Nulla id nisi enim. Sed tincidunt libero eu enim ultrices, at sagittis erat tempus. Nullam et dapibus lacus, sit amet fringilla augue. Aenean in sapien lorem.\r\n\r\nAenean accumsan orci in orci dapibus, eu viverra odio sodales. Donec odio tellus, blandit quis justo a, semper volutpat ante. Proin libero nisi, iaculis ac libero elementum, varius cursus tellus. Sed hendrerit finibus ipsum, non tristique urna maximus ac. Donec libero metus, feugiat ut mollis ut, pulvinar at mauris. Nunc quis facilisis sem. In nibh ligula, semper eget libero id, suscipit consectetur velit. Aenean in metus non tortor dictum dictum porta non augue. Vivamus eu risus id lorem consectetur lobortis vel in lacus. Aenean auctor blandit velit, ut semper urna pharetra id. Vivamus consequat quis lectus eu tristique. Vivamus vehicula tortor et leo dictum, vel bibendum risus porttitor. Etiam dictum malesuada orci sit amet euismod.\r\n\r\nIn non ante quis diam varius fringilla. In ornare at mi sed pretium. Ut quis est nec odio mattis imperdiet. Nullam et posuere elit, a ullamcorper lectus. Nulla sed consequat sapien, nec semper turpis. Fusce lorem ligula, fringilla non lectus eu, maximus porta orci. Nam efficitur porta ultricies. Vivamus ultrices lorem eu nisi aliquet, et condimentum nisl fermentum. Phasellus quis condimentum urna. Ut varius eleifend pharetra.\r\n\r\nSuspendisse dignissim scelerisque venenatis. Mauris felis est, sagittis at nibh at, tincidunt imperdiet justo. Mauris vulputate neque hendrerit arcu ornare blandit. Class aptent taciti sociosqu ad litora torquent per conubia nostra, per inceptos himenaeos. Nulla vel sollicitudin justo, ut dapibus nibh. Morbi quis cursus enim. Cras accumsan ultricies est at volutpat.",
"title": "Research programme and methodology",
"type": "2"
},
"15": {
"data": "In our lab we currently have results detailing:\r\n\r\nEtiam vehicula ultrices tortor vitae fringilla. Suspendisse cursus augue at risus venenatis ornare. Suspendisse cursus augue ac volutpat iaculis. Sed malesuada consequat commodo. Pellentesque nec enim sed ex ullamcorper fringilla in ac velit. Aenean vitae dolor vitae ante semper pharetra. Donec scelerisque ornare dolor sed congue. Fusce sed blandit diam.\r\n\r\nQuisque commodo nisl et lacus consequat, in euismod arcu porta. Pellentesque ac pellentesque urna, ut sollicitudin tellus. Cum sociis natoque penatibus et magnis dis parturient montes, nascetur ridiculus mus. Quisque leo lorem, tincidunt eu feugiat pretium, pellentesque et nunc. Curabitur commodo pharetra ante nec convallis. Sed at facilisis eros, vel fringilla ligula. Praesent hendrerit facilisis felis. Nunc iaculis mauris nec nunc pulvinar vehicula. Interdum et malesuada fames ac ante ipsum primis in faucibus. Aliquam viverra ipsum quis viverra pharetra. Ut quis varius felis.\r\n\r\nSed vel blandit ante. Donec sit amet dignissim nulla. Quisque tempus eu dui nec varius. In vehicula dui velit, ultricies vestibulum mi pharetra ac. Nunc nec imperdiet mi. Sed nec eros elementum, dictum risus sed, gravida purus. Suspendisse potenti. Aliquam congue gravida nisi, id consequat magna placerat ut. Ut non urna consectetur, congue est nec, interdum ex. Etiam venenatis mauris vitae velit iaculis mattis. Etiam vestibulum ipsum a dolor convallis condimentum. Mauris sed tortor ultrices, tristique nulla nec, blandit est. Vestibulum vitae ornare purus. Ut aliquet sapien non malesuada cursus.\r\n\r\nProin scelerisque aliquam sapien, ut vehicula eros. Integer sodales, leo eu imperdiet malesuada, eros ligula ultricies nisl, eget placerat nisi urna quis ante. Etiam venenatis massa et vestibulum pellentesque. Vivamus quis fringilla tellus. Phasellus id ullamcorper lacus, non convallis nisi. Ut vel risus odio. In porttitor blandit justo, a eleifend nulla scelerisque eu. Mauris non posuere sapien. Donec nulla elit, dapibus nec elit et, vehicula fringilla ligula. Sed eleifend pulvinar purus id pellentesque. Quisque lobortis vulputate nisl. Nam feugiat elit orci, quis consequat nunc lacinia quis.\r\n\r\nDonec feugiat nisi ac dui accumsan, sed sagittis velit rhoncus. Etiam ullamcorper risus vitae tortor pretium dapibus. Donec vehicula et erat ac venenatis. Etiam eu nisl sed augue vestibulum faucibus. Donec semper nisl nisl, quis semper mi dictum eu. Nunc dapibus felis sit amet lorem molestie, id blandit felis accumsan. Proin porttitor tristique purus non tempus. Duis ut cursus lorem. Maecenas sit amet ante nec ligula iaculis sodales ut interdum tortor. Integer diam felis, pharetra non est eu, ultricies condimentum eros. Phasellus erat lectus, tristique elementum est vitae, accumsan eleifend sem.",
"title": "Background in your lab & current results",
"type": "2"
},
"17": {
"data": "https:\/\/www.structuralbiology.eu\/update\/upload\/OfLyrce.png",
"title": "Files & figures (images & PDFs accepted)",
"type": "8"
},
"16": {
"data": "26811337,26698068,26711332",
"title": "Relevant publications",
"type": "7"
},
"42": {
"data": "{\"1872\":{\"microfocus\":\"1\",\"longwave\":\"0\",\"shifts\":\"3\",\"remote\":\"66\"}}",
"title": "iNEXT MX\/SAXS Specific",
"type": "0"
}
},
"visits": {
"1871": {
"vid": "1871",
"cid": "25",
"cpid": "595",
"plid": "171",
"acid": "18",
"center": "STRUBI",
"platform": "Crystallisation",
"centerplatform": "Nanodrop plate crystallization and imaging",
"access": "API Test Route"
},
"1872": {
"vid": "1872",
"cid": "290",
"cpid": "5040",
"plid": "283",
"acid": "18",
"center": "Diamond Light Source",
"platform": "X-ray crystallography beamline",
"centerplatform": "MX - Diamond",
"access": "API Test Route"
}
}
}
}
XML Data format is being maintained as a legacy format. Should the demand for XML return it will be documented here.
| Field ID | Field Name | Access Routes | Format | Output | Additional information |
|---|---|---|---|---|---|
| 11 | Scientific background and significance | Instruct Other/Collaboration FRISBI BioStructX iNEXT NMR ES iNEXT NMR HEDC iNEXT EM HEDC iNEXT SA iNEXT EM ES iNEXT MX/SAXS ES iNEXT SAXS SA iNEXT MX/SAXS HEDC iNEXT Mm Interactions API Test Route |
Long Text | Text | |
| 12 | Research programme and methodology | Instruct Other/Collaboration iNEXT EM HEDC iNEXT EM ES iNEXT Mm Interactions API Test Route |
Long Text | Text | |
| 13 | Ethical concerns | Instruct Other/Collaboration |
Yes/No if No Long Text | Text, blank if Yes | |
| 14 | Safety concerns | Instruct Other/Collaboration |
Yes/No if No Long Text | Text, blank if yes | |
| 15 | Background in your lab & current results | Instruct Other/Collaboration FRISBI BioStructX iNEXT NMR ES iNEXT NMR HEDC iNEXT EM HEDC iNEXT SA iNEXT EM ES iNEXT MX/SAXS ES iNEXT SAXS SA iNEXT MX/SAXS HEDC iNEXT Mm Interactions API Test Route |
Long Text | Text | |
| 16 | Relevant publications | Instruct Other/Collaboration FRISBI BioStructX iNEXT NMR ES iNEXT NMR HEDC iNEXT EM HEDC iNEXT SA iNEXT EM ES iNEXT MX/SAXS ES iNEXT SAXS SA iNEXT MX/SAXS HEDC iNEXT Mm Interactions API Test Route |
Publication List | 125232,1523223,161232 | A list of pubmed IDs comma delimited |
| 17 | Files & figures (images & PDFs accepted) | Instruct Other/Collaboration FRISBI BioStructX iNEXT NMR ES iNEXT NMR HEDC iNEXT EM HEDC iNEXT EM ES iNEXT MX/SAXS HEDC API Test Route |
File List | fileurl.jpg,fileurl.pdf | A list of full file URLs comma delimted |
| 19 | Funding | FRISBI | Option List | Selected option text | |
| 20 | Estimated access units | BioStructX | Short Text | Text | |
| 21 | 4 key words | BioStructX | Short Text | Text | |
| 22 | Value for translational research | iNEXT NMR ES iNEXT NMR HEDC iNEXT EM HEDC iNEXT SA iNEXT EM ES iNEXT MX/SAXS HEDC iNEXT Mm Interactions |
Long Text | Text | |
| 23 | Has the host been contacted? | iNEXT NMR ES iNEXT NMR HEDC |
Yes/No | yes/no | |
| 24 | Host collaborator | iNEXT NMR ES iNEXT NMR HEDC |
Short Text | Text | |
| 25 | Requested support | iNEXT NMR ES iNEXT NMR HEDC |
Option List | Selected option text | |
| 26 | Sample characteristics | iNEXT NMR ES iNEXT NMR HEDC |
Short Text | Text | |
| 27 | Has previous NMR work been performed? | iNEXT NMR ES iNEXT NMR HEDC |
Yes/No if No Long Text | Text, blank if yes | |
| 28 | Temperature for NMR experiments (°C) | iNEXT NMR ES iNEXT NMR HEDC |
Number | 37 | |
| 29 | Desired experiments | iNEXT NMR ES iNEXT NMR HEDC |
Short Text | Text | |
| 30 | Estimated measurement time | iNEXT NMR ES iNEXT NMR HEDC |
Short Text | Text | |
| 31 | Sequence of target(s) | iNEXT SA | Long Text | MANLCGWMLVLFVAT | |
| 32 | I can make 300ul of sample at concentration more than 5mg/ml | iNEXT SA | Yes/No | yes/no | |
| 33 | MW of (complex) macromolecule | iNEXT SA | Short Text | Text | |
| 34 | Gel of purified protein | iNEXT SA | File List | fileurl.jpg,fileurl.pdf | |
| 35 | (MW<40,000) I will provide 15N labeled sample (500ul, 0.2 mM) for HSQC/NMR | iNEXT SA | Yes/No | yes/no | |
| 36 | (MW>200,000) I will provide a sample (50 ul, 1mg/ml) for EM megative-stain analysis | iNEXT SA | Yes/No | yes/no | |
| 37 | Please upload an image of your previous cryoEM work | iNEXT EM HEDC | File List | ||
| 38 | Is your sample biosafety level 1 | iNEXT EM HEDC | Yes/No | yes/no | |
| 39 | Experimental plan | iNEXT EM ES | Long Text | Text | |
| 40 | Do you have an industrial collaboration or contract | iNEXT MX/SAXS HEDC | Yes/No | yes/no | |
| 41 | Do any of the envisaged samples have biosafety issues | iNEXT MX/SAXS HEDC | Yes/No | yes/no | |
| 42 | iNEXT MX/SAXS individual platform data | iNEXT MX/SAXS HEDC API Test Route |
Custom: inextmxsaxs | to be added | A JSON object with visit IDs containing an array of data relating to the visit, specific to MX or SAXS |
FAQ
Begin typing your question. If we don't have an answer for it in our FAQ, please leave us a message on our contact page.
-
How do I extract iNEXT MX/SAXS visit specific data?
To access the data within the iNEXT MX/SAXS custom field, you must first parse the content of FIELD ID 42 data as JSON into a native array. The JSON array is presented with keys that relate to the visit IDs. So first you need to traverse the array of visits associated with the proposal, find the one(s) that is accessing your centre, then use the visit ID to access the extra data held within field 42.

